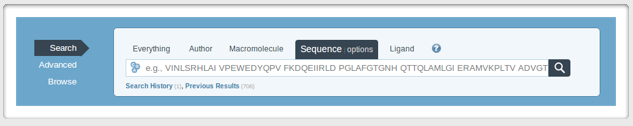
| If this still does not work, or you are getting a structure for only one portion of your
protein, it is possible that nothing similar was ever crystallized. Sometimes it is possible to
model
the structure of (a part of) your protein.
|
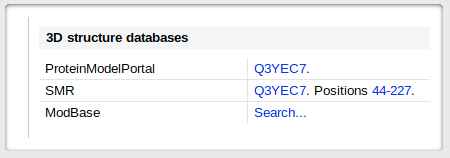 |
UNIPROT pages usually contain several links to modeling servers and databases. Here is an
arbitrary example - once on the page corresponding to
your protein, scroll down to the "Cross-references" section. You should see a small menu for the 3D structure databases,
like the one here, on the left. |
|
| |
|
|
| Still no results? Consider the possibility that your protein is
intrinsically disordered.
Disopred
is one of the websites that will give you a prediction, an estimate of how likely it
it is that your protein, or regions thereof, are disordered.
In this case we'll have to stick to 1D analysis. Cube will produce an xls spreadsheet and a png map
for that purpose.
|