| Since we want a reliable alignment, rather than remote homologues of our guery sequence,
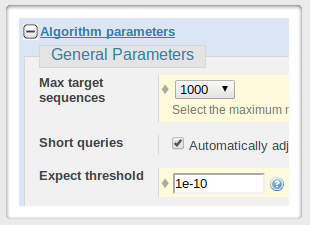
you might want to set the similarity ('Expect threshold') for your search to some small value.
You can also increase the number of sequences to be returned.
|  |
| Finally, select all sequences, and from Download menu select "FASTA (complete sequence)."
You might want to rename the file to something more descriptive then the name that NCBI's server provides.
|
| |
|
|
| An important caveat.
Getting sequences by simply blast-ing against a database is a strategy that will work for proteins
(or their coding genes) that have no close relatives
(paralogues)
in the same genome. The
worked example for the conservation case
is in that sense rigged: there, we are using a
protein without known paralogues in human and other vertebrates.
In the cases when this is not true - when your protein of interest falls in a whole family of related genes,
a different, safer, approach is needed. |